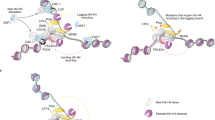
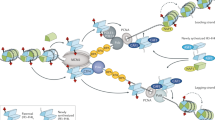
Key Points
-
Epigenetic inheritance concerns the mechanisms that ensure transmission of epigenetic marks from mother to daughter cell. Chromatin modifications and nuclear organization are candidates for epigenetic marks — whether they fulfil the criterion of heritability and what mechanisms ensure their propagation is an area of intensive research.
-
The passage of the replication fork challenges genetic and epigenetic information. Depending on the nature of the epigenetic mark, its inheritance can be ensured in a replication-coupled manner or in a timely manner that is separated from the disruptive event.
-
DNA methylation is inherited at the replication fork in a semi-conservative manner. The redistribution of parental histones, together with their parental modifications at the fork, affects the transmission of information. Histone chaperones have an important role in controlling histone dynamics at the fork. The maintenance of DNA methylation and histone modifications is interconnected at the replication fork.
-
The centromere-specific histone H3 variant CenH3 (CENP-A in humans) is inherited in a replication-uncoupled manner — new CENP-A is deposited in late telophase–G1 phase, highlighting a new window of inheritance during the cell cycle.
-
Restoration of pericentric heterochromatin after passage of the replication fork involves an RNA interference-dependent mechanism in fission yeast, whereas for mammals evidence suggests that the DNA methylation maintenance machinery, chromatin assembly factors and histone modifiers operate in a concerted manner to ensure heterochromatin maintenance.
-
Reprogramming during development highlights the reversibility of epigenetic states.
Abstract
Studies that concern the mechanism of DNA replication have provided a major framework for understanding genetic transmission through multiple cell cycles. Recent work has begun to gain insight into possible means to ensure the stable transmission of information beyond just DNA, and has led to the concept of epigenetic inheritance. Considering chromatin-based information, key candidates have arisen as epigenetic marks, including DNA and histone modifications, histone variants, non-histone chromatin proteins, nuclear RNA as well as higher-order chromatin organization. Understanding the dynamics and stability of these marks through the cell cycle is crucial in maintaining a given chromatin state.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Riggs, A. D., Martiennssen, R. A. & Russo, V. E. A. in Epigenetic Mechanisms of Gene Regulation 1–4 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1996).
Turner, B. M. Cellular memory and the histone code. Cell 111, 285–291 (2002).
Haig, D. The (dual) origin of epigenetics. Cold Spring Harb. Symp. Quant. Biol. 69, 67–70 (2004).
Ptashne, M. On the use of the word 'epigenetic'. Curr. Biol. 17, R233–R236 (2007).
Bird, A. Perceptions of epigenetics. Nature 447, 396–398 (2007).
Ledford, H. Language: disputed definitions. Nature 455, 1023–1028 (2008).
Waddington, C. H. The epigenotype. Endeavour 1 18–20 (1942).
Holliday, R. The inheritance of epigenetic defects. Science 238, 163–170 (1987).
Henikoff, S., Ahmad, K. & Malik, H. S. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102 (2001).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Loyola, A. & Almouzni, G. Marking histone H3 variants: how, when and why? Trends Biochem. Sci. 32, 425–433 (2007).
Corpet, A. & Almouzni, G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 19, 29–41 (2008).
Groth, A., Rocha, W., Verreault, A. & Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 128, 721–733 (2007).
Watson, J. D. & Crick, F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953).
DePamphilis, M. L. (ed.) DNA Replication and Human Disease (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2006).
Kunkel, T. A. & Burgers, P. M. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18, 521–527 (2008).
Moldovan, G. L., Pfander, B. & Jentsch, S. PCNA, the maestro of the replication fork. Cell 129, 665–679 (2007).
Alberts, B. Molecular Biology of the Cell (Garland Science Publishing, London, 2007).
Zhang, Z., Shibahara, K. & Stillman, B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408, 221–225 (2000).
Henderson, D. S., Banga, S. S., Grigliatti, T. A. & Boyd, J. B. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO J. 13, 1450–1459 (1994).
Shibahara, K. & Stillman, B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 (1999).
Moggs, J. G. et al. A CAF-1–PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20, 1206–1218 (2000).
Groth, A. et al. Regulation of replication fork progression through histone supply and demand. Science 318, 1928–1931 (2007). Shows that the histone H3 chaperone ASF1 exists in a complex with the putative replicative helicase and suggests that ASF1 handles both parental and new histones at the replication fork.
Bestor, T. H. & Ingram, V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl Acad. Sci. USA 80, 5559–5563 (1983).
Hermann, A., Goyal, R. & Jeltsch, A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 279, 48350–48359 (2004).
Chuang, L. S. et al. Human DNA-(cytosine-5) methyltransferase–PCNA complex as a target for p21WAF1. Science 277, 1996–2000 (1997).
Pradhan, S., Bacolla, A., Wells, R. D. & Roberts, R. J. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 274, 33002–33010 (1999).
Spada, F. et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 176, 565–571 (2007).
Schermelleh, L. et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 35, 4301–4312 (2007).
Woo, H. R., Pontes, O., Pikaard, C. S. & Richards, E. J. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21, 267–277 (2007).
Unoki, M., Nishidate, T. & Nakamura, Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 23, 7601–7610 (2004).
Sharif, J. et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 (2007).
Bostick, M. et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 (2007). The work reported in references 30, 32 and 33 identified the SRA-domain-containing protein NP95 and its homologue in A. thaliana as essential factors that bind to hemimethylated DNA and are required for faithful DNA methylation inheritance.
Arita, K., Ariyoshi, M., Tochio, H., Nakamura, Y. & Shirakawa, M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821 (2008).
Avvakumov, G. V. et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455, 822–825 (2008).
Hashimoto, H. et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455, 826–829 (2008).
Li, E., Bestor, T. H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
Jeddeloh, J. A., Stokes, T. L. & Richards, E. J. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nature Genet. 22, 94–97 (1999).
Brzeski, J. & Jerzmanowski, A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 278, 823–828 (2003).
Dennis, K., Fan, T., Geiman, T., Yan, Q. & Muegge, K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15, 2940–2944 (2001).
Zhang, F., Pomerantz, J. H., Sen, G., Palermo, A. T. & Blau, H. M. Active tissue-specific DNA demethylation conferred by somatic cell nuclei in stable heterokaryons. Proc. Natl Acad. Sci. USA 104, 4395–4400 (2007).
Metivier, R. et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452, 45–50 (2008).
Kangaspeska, S. et al. Transient cyclical methylation of promoter DNA. Nature 452, 112–115 (2008).
Gruenbaum, Y., Cedar, H. & Razin, A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature 295, 620–622 (1982).
Kimura, H. & Cook, P. R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153, 1341–1353 (2001).
Jackson, V. & Chalkley, R. A reevaluation of new histone deposition on replicating chromatin. J. Biol. Chem. 256, 5095–5103 (1981).
Polo, S. E. & Almouzni, G. Chromatin assembly: a basic recipe with various flavours. Curr. Opin. Genet. Dev. 16, 104–111 (2006).
De Koning, L., Corpet, A., Haber, J. E. & Almouzni, G. Histone chaperones: an escort network regulating histone traffic. Nature Struct. Mol. Biol. 14, 997–1007 (2007).
Stillman, B. Chromatin assembly during SV40 DNA replication in vitro. Cell 45, 555–565 (1986). The first report of chromatin assembly coupled in vitro to DNA replication.
Smith, S. & Stillman, B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58, 15–25 (1989).
Gaillard, P. H. et al. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell 86, 887–896 (1996).
Polo, S. E., Roche, D. & Almouzni, G. New histone incorporation marks sites of UV repair in human cells. Cell 127, 481–493 (2006).
Mello, J. A. et al. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3, 329–334 (2002).
Sobel, R. E., Cook, R. G., Perry, C. A., Annunziato, A. T. & Allis, C. D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl Acad. Sci. USA 92, 1237–1241 (1995).
Loyola, A., Bonaldi, T., Roche, D., Imhof, A. & Almouzni, G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24, 309–316 (2006).
Masumoto, H., Hawke, D., Kobayashi, R. & Verreault, A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 (2005).
Li, Q. et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134, 244–255 (2008).
Garcia, B. A. et al. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 282, 7641–7655 (2007).
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004). Shows that the histone variants H3.1 and H3.3 are assembled into chromatin by distinct histone chaperones and suggests that H3 and H4 are deposited as dimers.
Baxevanis, A. D., Godfrey, J. E. & Moudrianakis, E. N. Associative behavior of the histone (H3-H4)2 tetramer: dependence on ionic environment. Biochemistry 30, 8817–8823 (1991).
Verreault, A., Kaufman, P. D., Kobayashi, R. & Stillman, B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87, 95–104 (1996).
English, C. M., Adkins, M. W., Carson, J. J., Churchill, M. E. & Tyler, J. K. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006).
Natsume, R. et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341 (2007).
Song, J. J., Garlick, J. D. & Kingston, R. E. Structural basis of histone H4 recognition by p55. Genes Dev. 22, 1313–1318 (2008).
Murzina, N. V. et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16, 1077–1085 (2008).
Taverna, S. D., Li, H., Ruthenburg, A. J., Allis, C. D. & Patel, D. J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature Struct. Mol. Biol. 14, 1025–1040 (2007).
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).
Aagaard, L. et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18, 1923–1938 (1999).
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001). References 69 and 70 show that HP1 (Swi6 in fission yeast) binds to methylated H3K9 through its chromodomain and suggest that a self-perpetuating loop contributes to HP1 maintenance.
Hansen, K. H. et al. A model for transmission of the H3K27me3 epigenetic mark. Nature Cell Biol. 10, 1291–1300 (2008).
Leffak, I. M., Grainger, R. & Weintraub, H. Conservative assembly and segregation of nucleosomal histones. Cell 12, 837–845 (1977).
Milutinovic, S., Zhuang, Q. & Szyf, M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J. Biol. Chem. 277, 20974–20978 (2002).
Huen, M. S., Sy, S. M., van Deursen, J. M. & Chen, J. Direct interaction between SET8 and PCNA couples H4-K20 methylation with DNA replication. J. Biol. Chem. 283, 11073–11077 (2008).
Jorgensen, S. et al. The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 179, 1337–1345 (2007).
Poot, R. A. et al. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nature Cell Biol. 6, 1236–1244 (2004).
Sporbert, A., Gahl, A., Ankerhold, R., Leonhardt, H. & Cardoso, M. C. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell 10, 1355–1365 (2002).
Taddei, A., Roche, D., Sibarita, J. B., Turner, B. M. & Almouzni, G. Duplication and maintenance of heterochromatin domains. J. Cell Biol. 147, 1153–1166 (1999).
Sarraf, S. A. & Stancheva, I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 15, 595–605 (2004).
Reese, B. E., Bachman, K. E., Baylin, S. B. & Rountree, M. R. The methyl-CpG binding protein MBD1 interacts with the p150 subunit of chromatin assembly factor 1. Mol. Cell. Biol. 23, 3226–3236 (2003).
Fuks, F., Burgers, W. A., Brehm, A., Hughes-Davies, L. & Kouzarides, T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nature Genet. 24, 88–91 (2000).
Rountree, M. R., Bachman, K. E. & Baylin, S. B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genet. 25, 269–277 (2000).
Esteve, P. O. et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 20, 3089–3103 (2006).
Citterio, E. et al. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 24, 2526–2535 (2004).
Karagianni, P., Amazit, L., Qin, J. & Wong, J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 28, 705–717 (2008).
Ng, R. K. & Gurdon, J. B. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nature Cell Biol. 10, 102–109 (2008).
Zweidler, A. in Histone Genes: Structure, Organization and Regulation (eds Stein, G. S. et al.) 339–371 (Wiley, New York, 1984).
Henikoff, S., Furuyama, T. & Ahmad, K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 20, 320–326 (2004).
Ahmad, K. & Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 (2002).
McKittrick, E., Gafken, P. R., Ahmad, K. & Henikoff, S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA 101, 1525–1530 (2004).
Jin, C. & Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21, 1519–29 (2007).
Henikoff, S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nature Rev. Genet. 9, 15–26 (2008).
Ray-Gallet, D. et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 (2002).
Cleveland, D. W., Mao, Y. & Sullivan, K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 (2003).
Allshire, R. C. & Karpen, G. H. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nature Rev. Genet. 9, 923–937 (2008).
Hemmerich, P. et al. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180, 1101–1114 (2008).
Shelby, R. D., Monier, K. & Sullivan, K. F. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151, 1113–1118 (2000).
Jansen, L. E., Black, B. E., Foltz, D. R. & Cleveland, D. W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805 (2007). This elegant study, which uses SNAP-tag technology, shows that new CENP-A is deposited in a discrete time window at late telophase–G1 phase.
Sullivan, K. F. A solid foundation: functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11, 182–188 (2001).
Mizuguchi, G., Xiao, H., Wisniewski, J., Smith, M. M. & Wu, C. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell 129, 1153–1164 (2007).
Dalal, Y., Wang, H., Lindsay, S. & Henikoff, S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 5, e218 (2007).
Conde e Silva, N. et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 370, 555–573 (2007).
Bernard, P. et al. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542 (2001).
Nonaka, N. et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nature Cell Biol. 4, 89–93 (2002).
Yamada, T., Fischle, W., Sugiyama, T., Allis, C. D. & Grewal, S. I. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell 20, 173–185 (2005).
Chen, E. S. et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451, 734–737 (2008).
Kloc, A., Zaratiegui, M., Nora, E. & Martienssen, R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 18, 490–495 (2008). References 106 and 107 show that transcription and processing of centromeric repeats occurs in a discrete window during the cell cycle.
Kim, S. M., Dubey, D. D. & Huberman, J. A. Early-replicating heterochromatin. Genes Dev. 17, 330–335 (2003).
Volpe, T. A. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002). The first observation to link the RNAi pathway to heterochromatin maintenance in fission yeast.
Verdel, A. et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004).
Sugiyama, T., Cam, H., Verdel, A., Moazed, D. & Grewal, S.I. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl Acad. Sci. USA 102, 152–157 (2005).
Shankaranarayana, G. D., Motamedi, M. R., Moazed, D. & Grewal, S. I. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 13, 1240–1246 (2003).
Grewal, S. I. & Jia, S. Heterochromatin revisited. Nature Rev. Genet. 8, 35–46 (2007).
Fukagawa, T. et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nature Cell Biol. 6, 784–791 (2004).
Kanellopoulou, C. et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501 (2005).
Murchison, E. P., Partridge, J. F., Tam, O. H., Cheloufi, S. & Hannon, G. J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl Acad. Sci. USA 102, 12135–12140 (2005).
Rudert, F., Bronner, S., Garnier, J. M. & Dolle, P. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm. Genome 6, 76–83 (1995).
Lehnertz, B. et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 (2003).
Lu, J. & Gilbert, D. M. Cell cycle regulated transcription of heterochromatin in mammals vs. fission yeast: functional conservation or coincidence? Cell Cycle 7, 1907–1910 (2008).
Muchardt, C. et al. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 3, 975–981 (2002).
Maison, C. et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nature Genet. 30, 329–334 (2002).
Fischle, W. et al. Regulation of HP1–chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005).
Hirota, T., Lipp, J. J., Toh, B. H. & Peters, J. M. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438, 1176–1180 (2005).
Guenatri, M., Bailly, D., Maison, C. & Almouzni, G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166, 493–505 (2004).
Wu, R., Singh, P. B. & Gilbert, D. M. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J. Cell Biol. 174, 185–194 (2006).
Lu, J. & Gilbert, D. M. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J. Cell Biol. 179, 411–421 (2007).
Leonhardt, H., Page, A. W., Weier, H. U. & Bestor, T. H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71, 865–873 (1992).
Quivy, J. P. et al. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 23, 3516–3526 (2004).
Murzina, N., Verreault, A., Laue, E. & Stillman, B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4, 529–540 (1999).
Quivy, J. P., Gerard, A., Cook, A. J., Roche, D. & Almouzni, G. The HP1–p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nature Struct. Mol. Biol. 15, 972–979 (2008).
Houlard, M. et al. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2, e181 (2006).
Taddei, A., Maison, C., Roche, D. & Almouzni, G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nature Cell Biol. 3, 114–120 (2001).
Funabiki, H., Hagan, I., Uzawa, S. & Yanagida, M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121, 961–976 (1993).
Heitz, E. Das heterochromatin der moose. Jahrbuch Wiss Botanik, 762–818 (1928) (in German).
Brown, S. W. Heterochromatin. Science 151, 417–425 (1966).
Probst, A. V., Santos, F., Reik, W., Almouzni, G. & Dean, W. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma 116, 403–415 (2007).
Probst, A. V. & Almouzni, G. Pericentric heterochromatin: dynamic organization during early development in mammals. Differentiation 76, 15–23 (2008).
Mayer, W., Niveleau, A., Walter, J., Fundele, R. & Haaf, T. Demethylation of the zygotic paternal genome. Nature 403, 501–502 (2000). The first illustration of selective DNA demethylation of the paternal genome.
Santos, F., Hendrich, B., Reik, W. & Dean, W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241, 172–182 (2002).
Santos, F., Peters, A. H., Otte, A. P., Reik, W. & Dean, W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 280, 225–236 (2005).
Martin, C. et al. Genome restructuring in mouse embryos during reprogramming and early development. Dev. Biol. 292, 317–332 (2006).
Farthing, C. R. et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 4, e1000116 (2008).
Hajkova, P. et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452, 877–881 (2008).
Lee, J. et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 129, 1807–1817 (2002).
Hajkova, P. et al. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 117, 15–23 (2002).
Bannister, A. J. & Kouzarides, T. Reversing histone methylation. Nature 436, 1103–1106 (2005).
Rougier, N. et al. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 12, 2108–2113 (1998).
Howell, C. Y. et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104, 829–838 (2001).
Morgan, H. D., Santos, F., Green, K., Dean, W. & Reik, W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14, R47–R58 (2005).
Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 (2007).
Choi, Y. et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42 (2002).
Gong, Z. et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814 (2002).
Barreto, G. et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445, 671–675 (2007).
Jin, S. G., Guo, C. & Pfeifer, G. P. GADD45A does not promote DNA demethylation. PLoS Genet. 4, e1000013 (2008).
Ooi, S. K. & Bestor, T. H. The colorful history of active DNA demethylation. Cell 133, 1145–1148 (2008).
van der Heijden, G. W. et al. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev. Biol. 298, 458–469 (2006).
Hochedlinger, K. et al. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 18, 1875–1885 (2004).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). Demonstrates that somatic cells can be reprogrammed to an embryonic cell fate by forced expression of embryonic transcription factors.
Wernig, M. et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324 (2007).
Tada, M., Takahama, Y., Abe, K., Nakatsuji, N. & Tada, T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11, 1553–1558 (2001).
Roemer, I., Reik, W., Dean, W. & Klose, J. Epigenetic inheritance in the mouse. Curr. Biol. 7, 277–280 (1997).
Morgan, H. D., Sutherland, H. G., Martin, D. I. & Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nature Genet. 23, 314–318 (1999).
Rakyan, V. K. et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc. Natl Acad. Sci. USA 100, 2538–2543 (2003).
Buiting, K. et al. Epimutations in Prader–Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am. J. Hum. Genet. 72, 571–577 (2003).
Blewitt, M. E., Vickaryous, N. K., Paldi, A., Koseki, H. & Whitelaw, E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2, e49 (2006).
Cropley, J. E., Suter, C. M., Beckman, K. B. & Martin, D. I. Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation. Proc. Natl Acad. Sci. USA 103, 17308–17312 (2006).
Morgan, D. K. & Whitelaw, E. The case for transgenerational epigenetic inheritance in humans. Mamm. Genome 19, 394–397 (2008).
Lolle, S. J., Victor, J. L., Young, J. M. & Pruitt, R. E. Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434, 505–509 (2005).
Rassoulzadegan, M. et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441, 469–474 (2006).
Zacharioudakis, I., Gligoris, T. & Tzamarias, D. A yeast catabolic enzyme controls transcriptional memory. Curr. Biol. 17, 2041–2046 (2007).
Lepere, G., Betermier, M., Meyer, E. & Duharcourt, S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 22, 1501–1512 (2008).
Lippman, Z. & Martienssen, R. The role of RNA interference in heterochromatic silencing. Nature 431, 364–370 (2004).
McNairn, A. J. & Gilbert, D. M. Epigenomic replication: linking epigenetics to DNA replication. Bioessays 25, 647–656 (2003).
Fox, M. H., Arndt-Jovin, D. J., Jovin, T. M., Baumann, P. H. & Robert-Nicoud, M. Spatial and temporal distribution of DNA replication sites localized by immunofluorescence and confocal microscopy in mouse fibroblasts. J. Cell Sci. 99, 247–253 (1991).
O'Keefe, R. T., Henderson, S. C. & Spector, D. L. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 116, 1095–1110 (1992).
Hiratani, I. et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6, e245 (2008).
Gilbert, D. M. Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol. 14, 377–383 (2002).
Misteli, T. Beyond the sequence: cellular organization of genome function. Cell 128, 787–800 (2007).
Tatematsu, K. I., Yamazaki, T. & Ishikawa, F. MBD2–MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells 5, 677–688 (2000).
Bozhenok, L., Wade, P. A. & Varga-Weisz, P. WSTF–ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 21, 2231–2241 (2002).
Maison, C. & Almouzni, G. HP1 and the dynamics of heterochromatin maintenance. Nature Rev. Mol. Cell Biol. 5, 296–304 (2004).
van der Heijden, G. W. et al. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 122, 1008–1022 (2005).
Torres-Padilla, M. E., Bannister, A. J., Hurd, P. J., Kouzarides, T. & Zernicka-Goetz, M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50, 455–461 (2006).
Acknowledgements
We apologize for not quoting all of our colleagues for their contributions owing to space limitations. We thank D. Roche for providing images, J. P. Quivy and P. A. Defossez for critical comments to the manuscript. A.V.P. is supported by a European Molecular Biology Organization (EMBO) long-term fellowship. A.V.P., E.D. and G.A. are supported by La Ligue Nationale contre le Cancer (Equipe Labellisée la Ligue), Programme Incitatif Cooperatif (PIC) programmes 'Retinoblastome' and 'Replication, Instabilite chromosomique et cancer', the European Commission Network of Excellence Epigenome, ACI-2007-Cancéropôle IdF 'Breast cancer and Epigenetics' and the Agence Nationale de la Recherche.
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- Epigenetics
-
This term was coined by Waddington in 1942 to describe how genes of a genotype bring about a phenotype. Current definitions of epigenetics include the study of heritable changes in gene function that occur without alterations to the DNA sequence.
- Centromere
-
A region of a chromosome that is defined by the presence of a centromere-specific histone H3 variant (CenH3) and that functions as a platform for kinetochore assembly during mitosis.
- Heterochromatin
-
A chromatin region that remains condensed throughout the cell cycle and that is characterized by a specific chromatin signature.
- Reprogramming
-
The induced reversal of an epigenetic state, resulting in an altered cellular identity.
- Histone chaperone
-
A factor that associates with histones and stimulates a reaction that involves histone transfer without being part of the final product.
- DNA methyltransferase
-
An enzyme that transfers methyl groups from S-adenosylmethionine to specific adenines or cytosines in DNA.
- Histone H3 variant
-
A replicative histone H3 variant is expressed and incorporated during DNA replication (for example, H3.1 and H3.2), whereas a replacement variant is expressed throughout the cell cycle and is incorporated in a DNA-synthesis-independent manner (for example, H3.3 and the centromere-specific histone H3 variant CenH3).
- Histone deacetylase
-
An enzyme that removes acetyl groups from histones.
- Lys methyltransferase
-
An enzyme that catalyses the addition of a methyl group to specific Lys residues in histones and other non-histone proteins.
- Lys acetyltransferase
-
An enzyme that catalyses the addition of an acetyl group to specific Lys residues in histones and other non-histone proteins.
- Heterochromatin protein 1
-
(HP1). A chromodomain-containing protein that binds to methylated K9 on histone H3 and is associated with heterochromatin in fission yeast (Swi6), mammals (HP1) and Drosophila melanogaster (HP1).
- Pericentric heterochromatin
-
A heterochromatic region adjacent to chromatin containing the centromere-specific histone H3 variant CenH3, and which is considered to be typical constitutive heterochromatin.
- Small interfering RNA
-
A short, non-coding RNA (∼22-nt long) that is processed from longer double-stranded RNA by the RNA interference machinery. Such non-coding RNAs confer target specificity to the silencing complexes in which they reside.
- Phospho–methyl switch
-
The phosphorylation of histone H3S10 during late G2 phase and mitosis interferes with the binding of heterochromatin protein 1 to the adjacent methylated H3K9 residue.
- Chromocentre
-
A cluster of constitutive heterochromatin from different chromosomes that is formed during interphase.
Rights and permissions
About this article
Cite this article
Probst, A., Dunleavy, E. & Almouzni, G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10, 192–206 (2009). https://doi.org/10.1038/nrm2640
Issue Date:
DOI: https://doi.org/10.1038/nrm2640
This article is cited by
-
A latest progress in the study of fish behavior: cross-generational effects of behavior under pollution pressure and new technologies for behavior monitoring
Environmental Science and Pollution Research (2024)
-
The histone demethylase KDM5C functions as a tumor suppressor in AML by repression of bivalently marked immature genes
Leukemia (2023)
-
Comparative genomic analysis of the human genome and six bat genomes using unsupervised machine learning: Mb-level CpG and TFBS islands
BMC Genomics (2022)
-
Prebiotic synthesis and triphosphorylation of 3′-amino-TNA nucleosides
Nature Chemistry (2022)
-
Regulated interaction of ID2 with the anaphase-promoting complex links progression through mitosis with reactivation of cell-type-specific transcription
Nature Communications (2022)